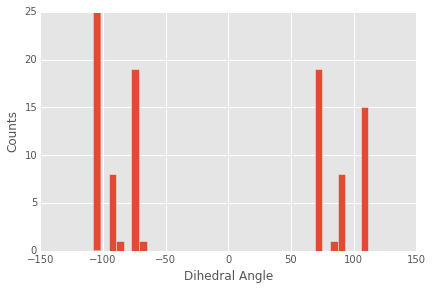
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem.Draw import MolDrawing, DrawingOptions
DrawingOptions.bondLineWidth=1.8
DrawingOptions.atomLabelFontSize=14
DrawingOptions.includeAtomNumbers=False
Create new molecule from a SMILES string:
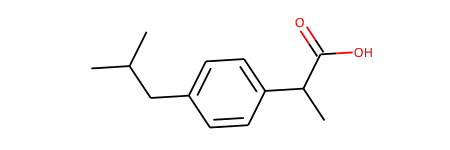
start_mol = Chem.MolFromSmiles('c1cc(CCCO ...